Summary Table (Download Table)
| Co-regulatory TF | Tissue | -log(P) |
  |   |   |
| PAX-2 | tongue | 6.41551 |
| OCT-1 | tongue | 7.31064 |
| LMO2_COMPLEX | tongue | 6.76922 |
| FAC1 | tongue | 6.57957 |
| E12 | tongue | 8.04858 |
| AREB6 | tongue | 7.40929 |
| GR | thymus | 10.0618 |
| ELK-1 | thymus | 6.39897 |
| XBP-1 | testis | 6.85089 |
| STAT6 | testis | 7.60769 |
| OCT-1 | testis | 7.70134 |
| GCNF | testis | 7.5514 |
| RSRFC4 | spleen | 7.40378 |
| LBP-1 | spleen | 8.20796 |
| GATA-2 | spleen | 6.37003 |
| AP-4 | spleen | 6.51605 |
| TCF-4 | soft_tissue | 6.19466 |
| LUN-1 | soft_tissue | 7.04451 |
| FOXO4 | soft_tissue | 8.19077 |
| DBP | soft_tissue | 6.732 |
| CHX10 | soft_tissue | 6.47967 |
| RSRFC4 | small_intestine | 6.674 |
| POU3F2 | small_intestine | 6.39512 |
| NKX6-2 | small_intestine | 9.50309 |
| NKX3A | small_intestine | 6.36779 |
| LHX3 | small_intestine | 15.3025 |
| GCNF | small_intestine | 7.07245 |
| AREB6 | skin | 6.53143 |
| LHX3 | placenta | 12.9153 |
| ATF | placenta | 6.94674 |
| TBP | peripheral_nervous_system | 11.8041 |
| POU3F2 | peripheral_nervous_system | 10.8939 |
| OCT-1 | peripheral_nervous_system | 9.21086 |
| NKX3A | peripheral_nervous_system | 8.98791 |
| NF-Y | peripheral_nervous_system | 6.16275 |
| IPF1 | peripheral_nervous_system | 7.99739 |
| HNF-3ALPHA | peripheral_nervous_system | 9.08456 |
| HFH-3 | peripheral_nervous_system | 8.7821 |
| FREAC-7 | peripheral_nervous_system | 6.43794 |
| CDP | peripheral_nervous_system | 7.44175 |
| CART-1 | peripheral_nervous_system | 6.72054 |
| C/EBP | peripheral_nervous_system | 6.67107 |
| ALX-4 | peripheral_nervous_system | 11.6025 |
| TAL-1BETA/E47 | pancreas | 6.56683 |
| PPARG | pancreas | 6.82133 |
| PPARALPHA/RXR-ALPHA | pancreas | 6.76899 |
| LHX3 | pancreas | 6.45407 |
| GATA-3 | pancreas | 6.79679 |
| E47 | pancreas | 12.21 |
| AREB6 | pancreas | 9.27013 |
| LXR | ovary | 6.73567 |
| USF | muscle | 8.76963 |
| TAL-1BETA/ITF-2 | muscle | 7.54634 |
| TAL-1BETA/E47 | muscle | 7.30926 |
| RSRFC4 | muscle | 9.11595 |
| MYOD | muscle | 6.90176 |
| MEF-2 | muscle | 14.8477 |
| MAX | muscle | 6.75544 |
| E4BP4 | muscle | 6.36414 |
| AP-4 | muscle | 6.46829 |
| RSRFC4 | mammary_gland | 7.06902 |
| FAC1 | mammary_gland | 8.2781 |
| PU.1 | lymph_node | 6.23369 |
| TITF1 | liver | 6.55621 |
| SREBP-1 | liver | 6.64997 |
| LMO2_COMPLEX | liver | 6.65199 |
| HNF-4ALPHA | liver | 9.35388 |
| HNF-4 | liver | 6.31596 |
| GATA-6 | liver | 7.34516 |
| GATA-1 | liver | 6.84441 |
| CRX | liver | 7.10247 |
| BACH1 | liver | 6.18678 |
| TFIIA | larynx | 7.27922 |
| PBX1B | larynx | 6.51671 |
| PAX-2 | larynx | 6.58782 |
| ISRE | larynx | 6.36869 |
| IRF1 | larynx | 7.09741 |
| ICSBP | larynx | 7.26881 |
| E47 | larynx | 8.56163 |
| STAT4 | kidney | 7.36854 |
| OCT-1 | kidney | 7.90319 |
| HNF-4ALPHA | kidney | 9.60021 |
| HNF-4 | kidney | 12.0581 |
| HNF-1 | kidney | 10.4516 |
| GATA-1 | kidney | 6.46025 |
| CDC5 | kidney | 8.20255 |
| TST-1 | heart | 8.89209 |
| TFIIA | heart | 7.49386 |
| TCF-4 | heart | 8.10651 |
| TAL-1BETA/E47 | heart | 6.2356 |
| TAL-1ALPHA/E47 | heart | 7.59047 |
| STAT4 | heart | 10.1672 |
| SRF | heart | 8.57008 |
| RSRFC4 | heart | 7.0458 |
| POU1F1 | heart | 6.74534 |
| PITX2 | heart | 16.97 |
| NKX6-2 | heart | 7.14547 |
| MEF-2 | heart | 19.4159 |
| LMO2_COMPLEX | heart | 9.87205 |
| HNF-1 | heart | 10.9916 |
| HEB | heart | 10.3071 |
| GATA-X | heart | 9.38028 |
| GATA-6 | heart | 13.1626 |
| GATA-1 | heart | 10.8074 |
| FXR | heart | 6.18204 |
| CDP | heart | 8.60131 |
| C-MYC/MAX | heart | 8.01682 |
| AP-1 | heart | 6.26132 |
| AMEF-2 | heart | 14.494 |
| AFP1 | heart | 10.5312 |
| SREBP-1 | eye | 7.91676 |
| NRSF | eye | 6.45537 |
| NF-Y | eye | 11.1534 |
| MYOGENIN/NF-1 | eye | 7.70954 |
| MEF-2 | eye | 6.64555 |
| CRX | eye | 7.06785 |
| CDP_CR3 | eye | 6.73801 |
| CDP | eye | 10.2419 |
| C/EBP | eye | 7.62129 |
| ARP-1 | eye | 7.21042 |
| ALPHA-CP1 | eye | 10.7371 |
| XBP-1 | brain | 7.17917 |
| TGIF | brain | 6.39591 |
| TEF | brain | 7.61382 |
| TCF-4 | brain | 12.3635 |
| TAL-1BETA/ITF-2 | brain | 6.48226 |
| SRY | brain | 6.23264 |
| SRF | brain | 6.86692 |
| RREB-1 | brain | 13.312 |
| PAX-1 | brain | 9.63826 |
| NRSF | brain | 9.70834 |
| MAZ | brain | 13.1692 |
| LUN-1 | brain | 6.56541 |
| LF-A1 | brain | 14.3153 |
| LEF-1 | brain | 10.2512 |
| IPF1 | brain | 7.20735 |
| GCM | brain | 11.1165 |
| GATA-4 | brain | 7.61643 |
| GATA-2 | brain | 8.11566 |
| FREAC-3 | brain | 8.86646 |
| CHOP-C/EBPALPHA | brain | 9.35397 |
| AP-4 | brain | 6.75125 |
| GR | bone_marrow | 6.27063 |
| RFX1 | bone | 20.7842 |
| MIF-1 | bone | 23.0105 |
| EF-C | bone | 17.9911 |
| IRF-1 | bladder | 6.62733 |
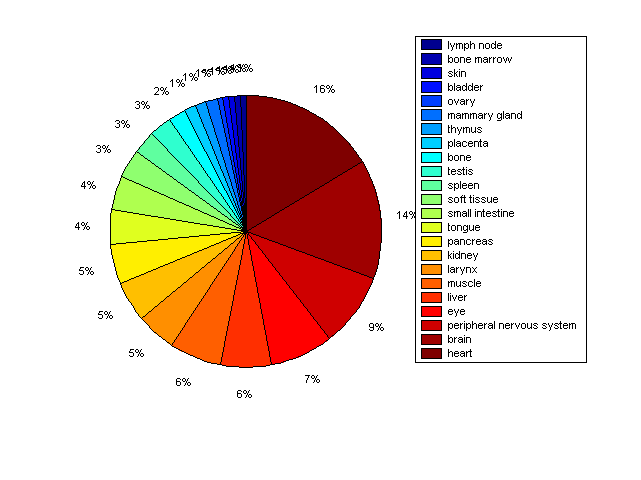
Distribution of Co-regulatory TFs
|
|
|
Description: this pie chart displays the distribution of co-regulatory transcription factors (TFs) in different tissues. Color schema: the tissue with the largest percentage of co-regulatory TFs is colored dark red whereas the tissue with the smallest percentage of co-regulatory TFs is colored dark blue. Tissues with intermediate percentages of co-regulatory TFs are colored from light red to yellow and cyan and then to light blue. |