Summary Table (Download Table)
| Co-regulatory TF | Tissue | -log(P) |
  |   |   |
| OCT-1 | bladder | 6.9093 |
| BACH2 | blood | 6.69071 |
| C/EBP | blood | 6.3906 |
| OCT-1 | blood | 6.16849 |
| OCT-1 | bone | 7.4895 |
| GR | bone_marrow | 7.86221 |
| CHOP-C/EBPALPHA | brain | 9.40541 |
| CREB | brain | 7.62396 |
| FREAC-3 | brain | 10.3001 |
| FREAC-7 | brain | 7.65459 |
| HNF-3ALPHA | brain | 11.2019 |
| LBP-1 | brain | 8.75482 |
| MEF-2 | brain | 7.21627 |
| MIF-1 | brain | 7.87062 |
| MRF-2 | brain | 8.72998 |
| NRL | brain | 7.426 |
| OSF2 | brain | 6.60756 |
| PBX-1 | brain | 7.01904 |
| RREB-1 | brain | 10.2585 |
| SOX-9 | brain | 9.85842 |
| TST-1 | brain | 13.3686 |
| XBP-1 | brain | 7.66777 |
| HNF-1 | colon | 6.96 |
| CRX | eye | 6.24893 |
| HSF2 | eye | 6.64985 |
| AMEF-2 | heart | 8.4579 |
| AP-1 | heart | 6.32125 |
| CDC5 | heart | 6.54654 |
| FXR | heart | 6.56962 |
| MEF-2 | heart | 10.5636 |
| NKX6-1 | heart | 6.71561 |
| RSRFC4 | heart | 13.9055 |
| TATA | heart | 9.5415 |
| TST-1 | heart | 7.03935 |
| NKX3A | kidney | 7.29621 |
| TATA | larynx | 7.65802 |
| ALPHA-CP1 | liver | 6.68364 |
| C/EBPGAMMA | liver | 11.7979 |
| FXR | liver | 6.8881 |
| HNF-1 | liver | 17.4039 |
| HNF-4 | liver | 6.53284 |
| HNF-4ALPHA | liver | 7.09048 |
| LMO2_COMPLEX | liver | 6.47625 |
| MEIS1B/HOXA9 | liver | 6.51494 |
| NR2E3 | liver | 7.1232 |
| NF-1 | lung | 6.94306 |
| LBP-1 | lymph_node | 7.11356 |
| LHX3 | lymph_node | 6.78464 |
| AP-4 | muscle | 11.4931 |
| BACH1 | muscle | 8.46914 |
| C-MYC/MAX | muscle | 6.27765 |
| C/EBP | muscle | 11.5802 |
| C/EBPBETA | muscle | 6.69932 |
| CDP | muscle | 12.9793 |
| CDP_CR1 | muscle | 6.70556 |
| ERR_ALPHA | muscle | 10.5925 |
| GATA-4 | muscle | 14.5253 |
| HEB | muscle | 15.5396 |
| HNF-4ALPHA | muscle | 10.4411 |
| HOXA4 | muscle | 8.37695 |
| IRF-1 | muscle | 6.73258 |
| MAX | muscle | 6.23417 |
| MEF-2 | muscle | 15.0957 |
| NR2E3 | muscle | 12.0096 |
| PPARG | muscle | 6.35817 |
| TATA | muscle | 12.1428 |
| TITF1 | muscle | 11.5081 |
| USF | muscle | 7.82397 |
| XBP-1 | muscle | 7.89394 |
| ATF | peripheral_nervous_system | 9.95285 |
| ATF3 | peripheral_nervous_system | 7.90894 |
| GCNF | peripheral_nervous_system | 9.16725 |
| HFH-4 | peripheral_nervous_system | 6.45005 |
| MAZR | peripheral_nervous_system | 10.1307 |
| POU3F2 | peripheral_nervous_system | 7.04549 |
| SRF | peripheral_nervous_system | 6.45924 |
| SRY | peripheral_nervous_system | 10.4152 |
| TEF | peripheral_nervous_system | 9.84909 |
| YY1 | peripheral_nervous_system | 6.81126 |
| C/EBPALPHA | placenta | 7.61575 |
| TATA | placenta | 6.38015 |
| LHX3 | prostate | 9.25791 |
| C-ETS-1 | soft_tissue | 6.41915 |
| C-ETS-2 | soft_tissue | 10.6773 |
| MEIS1A/HOXA9 | soft_tissue | 6.52042 |
| SRY | soft_tissue | 9.06506 |
| TATA | soft_tissue | 6.85457 |
| TCF11 | soft_tissue | 9.36401 |
| TEL-2 | soft_tissue | 6.45163 |
| TGIF | soft_tissue | 6.17291 |
| HNF-1 | spleen | 7.98276 |
| IRF-7 | tongue | 7.33886 |
| MRF-2 | tongue | 6.47935 |
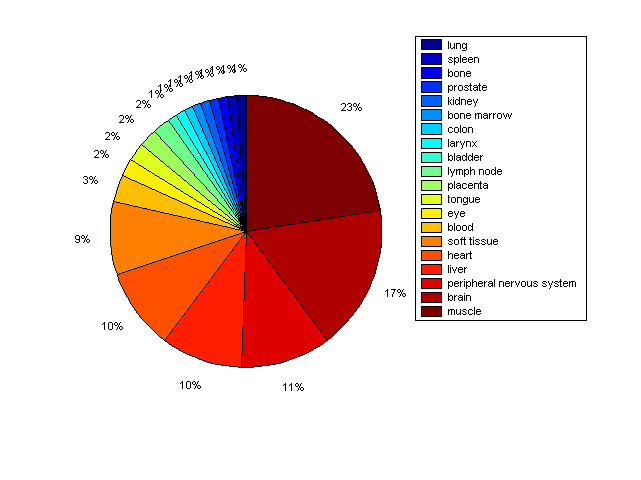
Distribution of Co-regulatory TFs
|
|
|
Description: this pie chart displays the distribution of co-regulatory transcription factors (TFs) in different tissues. Color schema: the tissue with the largest percentage of co-regulatory TFs is colored dark red whereas the tissue with the smallest percentage of co-regulatory TFs is colored dark blue. Tissues with intermediate percentages of co-regulatory TFs are colored from light red to yellow and cyan and then to light blue. |